Science fiction writer Arthur C. Clarke’s 3rd law states that “any sufficiently advanced technology is indistinguishable from magic.” Indika Rajapakse, Ph.D., is a believer. The engineer and mathematician now discovers himself a biologist. And he believes the charm of blending these 3 disciplines is crucial to unraveling how cells function.
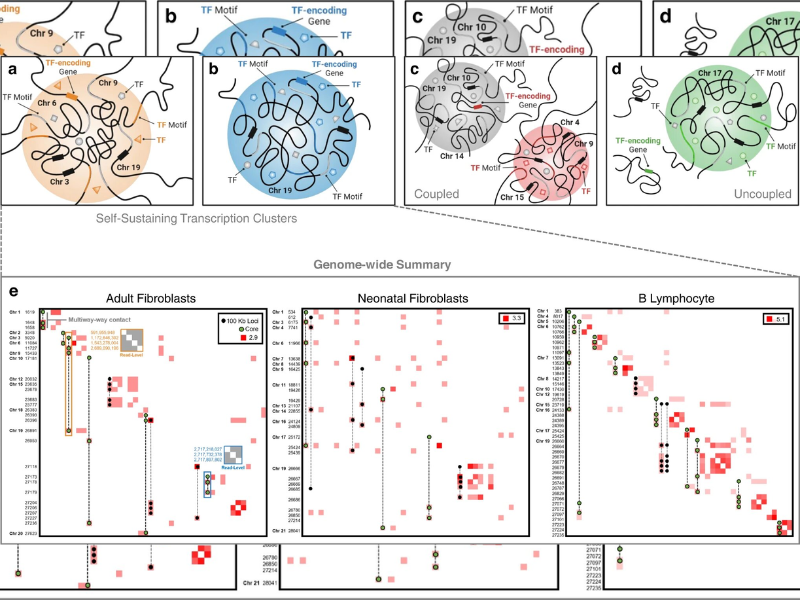
His latest development is a recent mathematical technique to start understanding how a cell’s nucleus is organized. The method, which Rajapakse and collaborators tested on several types of cells, revealed what the researchers termed self-sustaining transcription collections, a subset of proteins that play a crucial role in keeping cell identity.
They hope this comprehension will expose vulnerabilities that could be targeted to reprogram one cell to stop cancer or other illnesses.
“More and more cancer biologists believe genome organization plays a big role in understanding uncontrollable cell division and also whether we can reprogram a cancer cell.
That means we must understand more detail about what’s occurring in the nucleus,” stated Rajapakse, associate professor of computational medicine and bioinformatics, maths, and also biomedical engineering at the College of Michigan. He is also one member of the U-M Rogel Cancer Center.
Rajapakse is the senior writer on the paper, released in Nature Communications. The project was led by a trio of graduate students with an interdisciplinary group of researchers.
The group improved upon an older technology to study chromatin, called Hi-C, that maps which pieces of the genome are close together. It could identify chromosome translocations, like those that happen in some cancers. Its limitation, however, is that it observes only these adjacent genomic regions.
The recent technology, called Pore-C, uses much more information to visualize how all of the pieces within a cell’s nucleus interact. The researchers utilized a mathematical technique called hypergraphs. Think: three-dimensional Venn diagram. It allows scientists to see not simply pairs of genomic regions that interact but the totality of the complex and overlapping genome-wide connections within the cells.
“This multi-dimensional relationship we can understand unambiguously. It gives us a more straightforward way to understand organizational principles inside the nucleus. If you comprehend that, you can also understand where these organizational principles deviate, like in cancer,” Rajapakse stated. “This is like putting three worlds together– technology, mathematics, and biology– to analyze more detail inside the nucleus.”
The scientists tested their method on neonatal fibroblasts, biopsied adult fibroblasts, and B lymphocytes. They identified organizations of transcription clusters specific to each cell kind. They also discovered what they called self-sustaining transcription clusters, which serve as key transcriptional signatures for a cell type.
Rajapakse describes this as the first step in one bigger picture.
“My objective is to construct this type of picture over the cell cycle to understand how a cell goes through different stages. Cancer is uncontrollable cell division,” Rajapakse stated. “If we comprehend how a normal cell changes over time, we can begin to examine controlled, and uncontrolled systems and also find ways to reprogram that system.”
More information:
Gabrielle A. Dotson et al, Deciphering multi-way interactions in the human genome, Nature Communications (2022). DOI: 10.1038/s41467-022-32980-z
Read the original article on PHYS.