Bloodborne Pathogens Swiftly Identified by Melting their DNA

David Baillot/UC San Diego Jacobs School of Engineering
A new technique that melts bacterial DNA in blood samples could diagnose life-threatening infections faster than before.
Typically, to detect harmful bacteria in a person’s bloodstream, a blood sample is cultured in a Petri dish containing a growth medium. If bacteria are present, they will proliferate. However, it may take anywhere from 15 hours to several days for them to reach detectable levels.
Innovative Microfluidic Chip Design for Rapid Bacterial Detection
However, Under the guidance of Prof. Stephanie Fraley, researchers at the University of California San Diego have been investigating a faster and more precise solution. Moreover, They devised a microfluidic chip where a small blood sample is placed and heated to temperatures ranging from 50 to 90 ºC (122 to 194 ºF). If bacteria are present in the sample, the heat prompts their DNA molecules to melt. This melting leads to the unwinding of the double-helix strands in a distinct pattern unique to their nucleotide sequence.
In fact, “To identify this pattern, a specialized dye is introduced into the sample, causing the unwinding to emit fluorescent light. Analyzing the properties of this light yields a signature called a melting curve. This curve is then compared to known curves associated with specific bacteria. Upon finding a match, the bacteria in the blood sample are identified. The entire process is completed in under six hours.Custom machine-learning algorithms enable this speed by identifying and removing the patient’s DNA melting curve and other background noise.
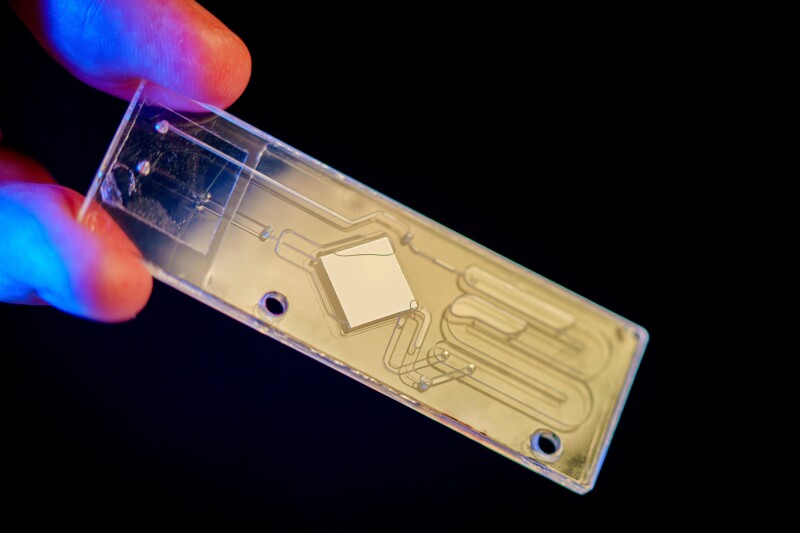
David Baillot/UC San Diego Jacobs School of Engineering
Comparative Evaluation of the New Technology on Suspected Sepsis Patients
To conclude, in an evaluation of the technology, blood samples from 17 children, suspected of having severe sepsis infections, were examined. However, the new technique yielded results identical to those obtained through conventional methods and did not yield any false positives. This contrasts with nucleic acid amplification methods, which amplify all DNA signatures indiscriminately, often leading to false positives. However, “this study marks the first application of this method to whole blood samples from suspected sepsis patients,” noted Fraley, emphasizing the study’s significance in simulating real clinical scenarios.
Read the original article on: New Atlas
Read more: Human-cell Biobots Boost Neuron Growth Sans DNA Mods











Comments (2)
[…] Read more: Bloodborne Pathogens Swiftly Identified by Melting their DNA […]
[…] Read more: Bloodborne Pathogens Swiftly Identified by Melting their DNA […]
Comments are closed.